Statement of Significance
Freshwater aquarium shrimp breeding relies on empirical knowledge about which species can interbreed, but quantitative predictions of compatibility remain limited. We used molecular phylogenetics to measure genetic distances between 17 popular aquarium shrimp species and calibrated a breeding compatibility scale using well-documented hybridization outcomes in felids. Phylogenetic distances within Caridina ranged from 0.12-1.50 substitutions/site, with only three species pairs falling below the 0.15 compatibility threshold. Caridina serrata (Tangerine Tiger) and C. cantonensis (Crystal Red) showed 0.39 genetic distance, exceeding the 0.25 incompatibility threshold. In contrast, all Neocaridina species pairs clustered within 0.06-0.11, indicating high interbreeding potential. This framework provides quantitative, evidence-based predictions of breeding compatibility for aquarium species management and selective breeding programs.
Methods
Sequence Acquisition
COI (cytochrome c oxidase subunit I) sequences for 17 freshwater aquarium shrimp species and 17 feline species were obtained from GenBank using an automated search pipeline. For each species, the pipeline prioritised complete mitochondrial genomes, followed by complete COI genes (≥1500 bp), then partial COI sequences (≥400 bp). Shrimp taxa included representatives from Caridina, Neocaridina, Atyopsis, Atya, Palaemon, and Macrobrachium, selected for commercial availability and phylogenetic coverage. Feline taxa spanned family Felidae plus two Feliformia outgroups (Crocuta crocuta, Cryptoprocta ferox) for calibration purposes. Final sequence lengths ranged from 400-1545 bp. Felids were selected for calibration due to well-documented hybridisation outcomes across a range of genetic distances, from fertile intraspecific crosses to reproductively incompatible intergeneric pairs.
Sequence Alignment and Phylogenetic Reconstruction
All 34 sequences (17 shrimp + 17 felids) were aligned using MAFFT via the NGPhylogeny.fr web portal. A single maximum likelihood phylogenetic tree was constructed using PhyML 3.3 with Smart Model Selection (SMS) for automatic model optimization based on the Akaike Information Criterion. The selected model was GTR+Γ+I (Gamma shape α = 0.389, 44.3% invariant sites). Branch support was assessed using approximate likelihood ratio tests. This combined tree enabled direct distance comparisons between shrimp and felids for calibration purposes. Clade-specific subtrees (shrimp-only and felid-only) were subsequently extracted from the unified tree for visualization.
Distance Analysis and Breeding Compatibility Calibration
Pairwise phylogenetic distances (substitutions/site) were calculated from the unified tree by summing branch lengths between terminal nodes. The combined tree was visualized as an unrooted phylogeny using iTOL (Interactive Tree of Life), and clade-specific subtrees (shrimp-only and felid-only) were extracted and rendered separately as unrooted trees to highlight within-group relationships. Distance matrices were visualized as clustered heatmaps using hierarchical clustering with a non-linear color scale (power transformation γ = 0.4) to emphasize variation at low distances where hybridization potential is highest.
A breeding compatibility scale was calibrated using documented feline hybridization outcomes where fertility is known: Felis catus × F. silvestris (distance 0.005, fertile), Panthera intrageneric crosses (0.07-0.14, viable with possible male sterility), and intergeneric felid pairs (>0.25, no documented hybrids). Thresholds were defined as: <0.05 (highly compatible), 0.05-0.15 (compatible), 0.15-0.25 (marginally compatible), >0.25 (incompatible). These thresholds were applied to predict shrimp interbreeding potential based on COI divergence.
Visualisations
Figure 1a. Unrooted phylogeny of 17 freshwater aquarium shrimp species.
Figure 1b. Unrooted phylogeny of 17 felid species and two Feliformia outgroups.
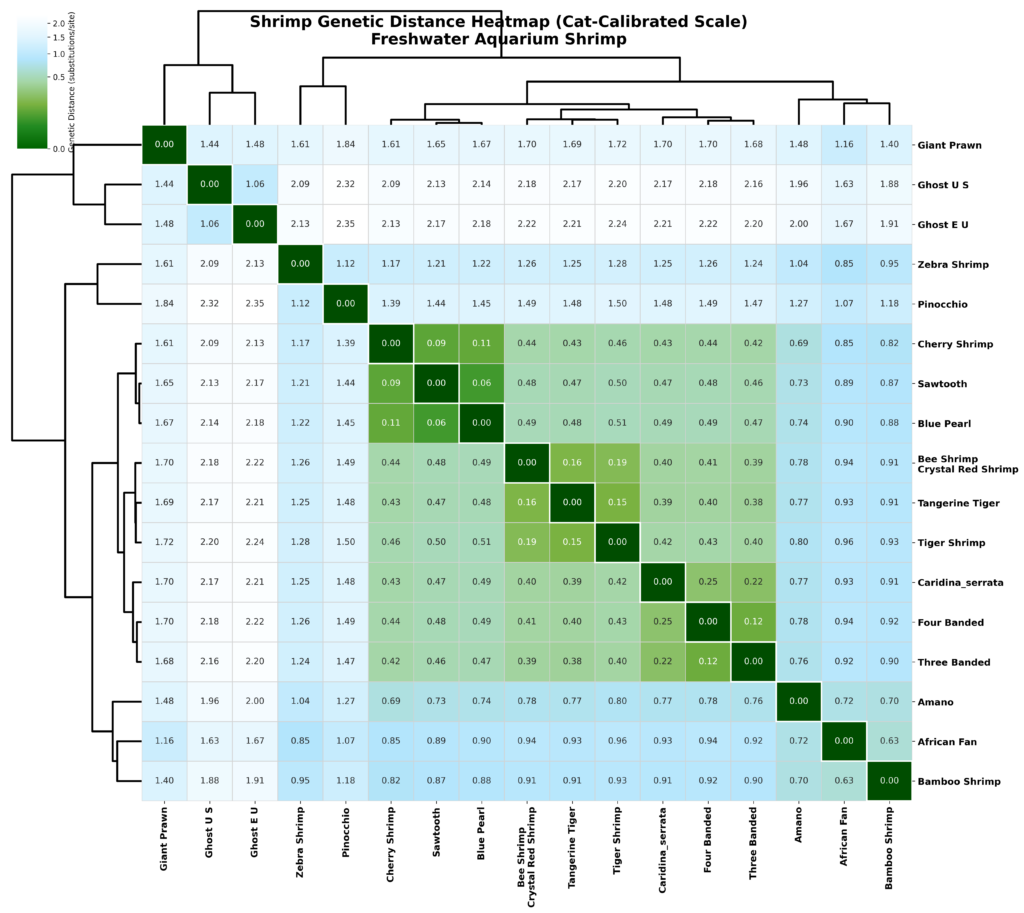
Figure 2. Pairwise phylogenetic distance heatmap for aquarium shrimp.
Phylogenetic Context and Interpretation
The phylogenetic topology recovered a monophyletic C. serrata species group with short internal branches and evidence of recent divergence, consistent with the rapid radiation documented by Klotz & von Rintelen (2014). Genetic distances within Caridina ranged from 0.12-1.50 substitutions/site, comparable to the 6-29% COI divergence observed within the Caridina clinata species complex by Hou et al. (2025), demonstrating that the genus encompasses substantial cryptic diversity. Using felid-calibrated breeding compatibility thresholds, three Caridina pairs fell within the compatible range (<0.15): C. tetrazona × C. trifasciata (0.12), C. cantonensis × C. mariae (0.15), and C. cantonensis × C. logemanni (0.16). The distance between C. serrata and C. cantonensis was 0.39, exceeding the 0.25 incompatibility threshold. Neocaridina species clustered tightly (0.06-0.11), within the compatible range for all pairwise comparisons.
References
Hou, J., Zhang, J., Chen, B., Zhang, Y., Chen, W., & Guo, Z. (2025). Integrative taxonomy reveals the Caridina clinata Cai, Nguyen & Ng, 1999 species complex (Crustacea, Decapoda, Atyidae), with descriptions of eight new species from Hainan Island, China. Zoosystematics and Evolution, 101(4), 2295-2336. https://doi.org/10.3897/zse.101.172207
Klotz, W., & von Rintelen, T. (2014). To “bee” or not to be—on some ornamental shrimp from Guangdong Province, Southern China and Hong Kong SAR, with descriptions of three new species. Zootaxa, 3889(2), 151-184. https://doi.org/10.11646/zootaxa.3889.2.1